Guest 10 - Bioinformatics
Lecture 27 - March 22nd, 2017
Guest Lecture
by Prof. Ulrike Stege
Some bioinformatics / Computational Biology: An algorithm to Build Evolutionary Trees
- What is bioinformatics / computational biology?
- An Interdiscipline field addressig bio
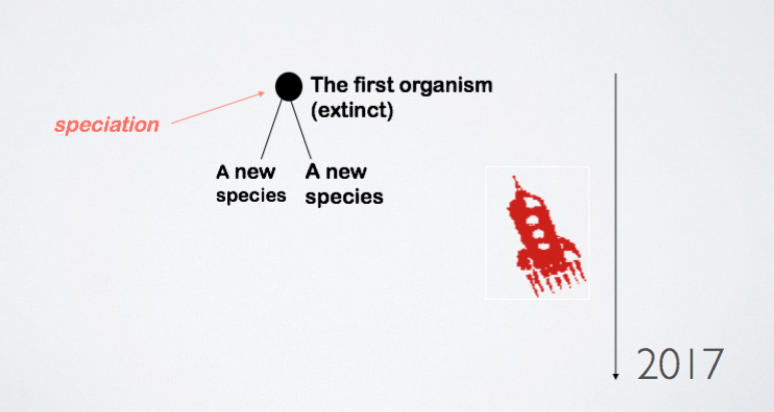
- Origin of species
- What does the tree of life look like?
- Sequencing of species/organismsn/viruses
- What evolutionary events do exist? When and how often did or do ther occur?
Evolutionary Trees (An Other Application of Graphs)
How did life evolve?

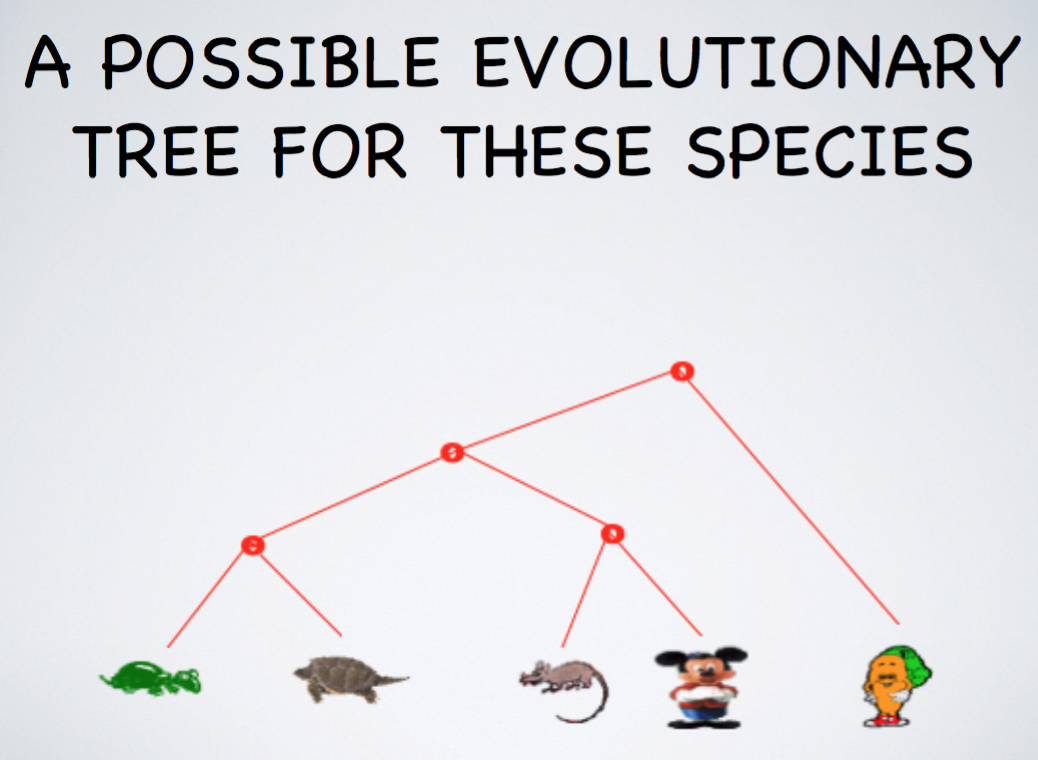
Determining Evolutionary Trees Today
- Of course, today we do not use looks only, rather consider more informative data such as the organisms's DNA sequences
- In addition, we might want to use other properties or characteristics known
- Today, we look at an algotritm that uses any type....
REMARK: trees are a special type of graphs (graphs without cycles)
- Acyclic graphs
- Consists of two types of vertices/nodes:
- External vertices (leaves)
- Internal vertices
Bug mutation: How did these bugs evolve???
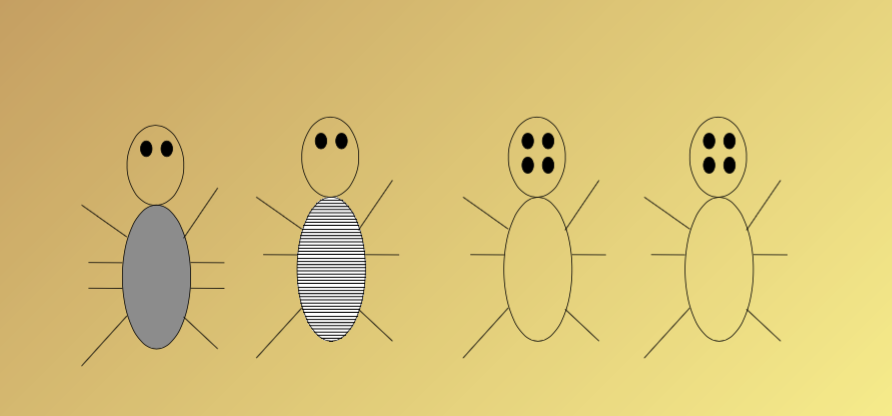
What did the ancestors of these bugs look like???
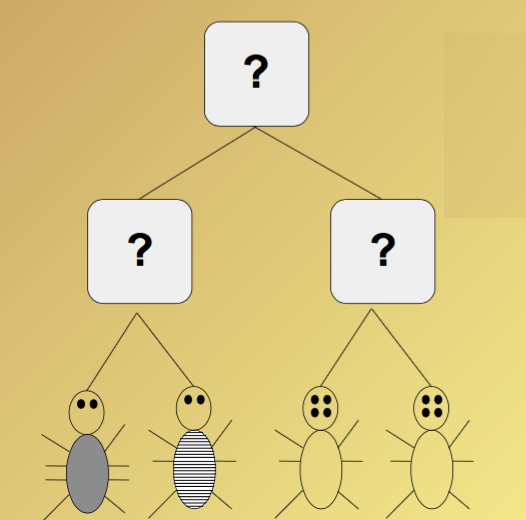
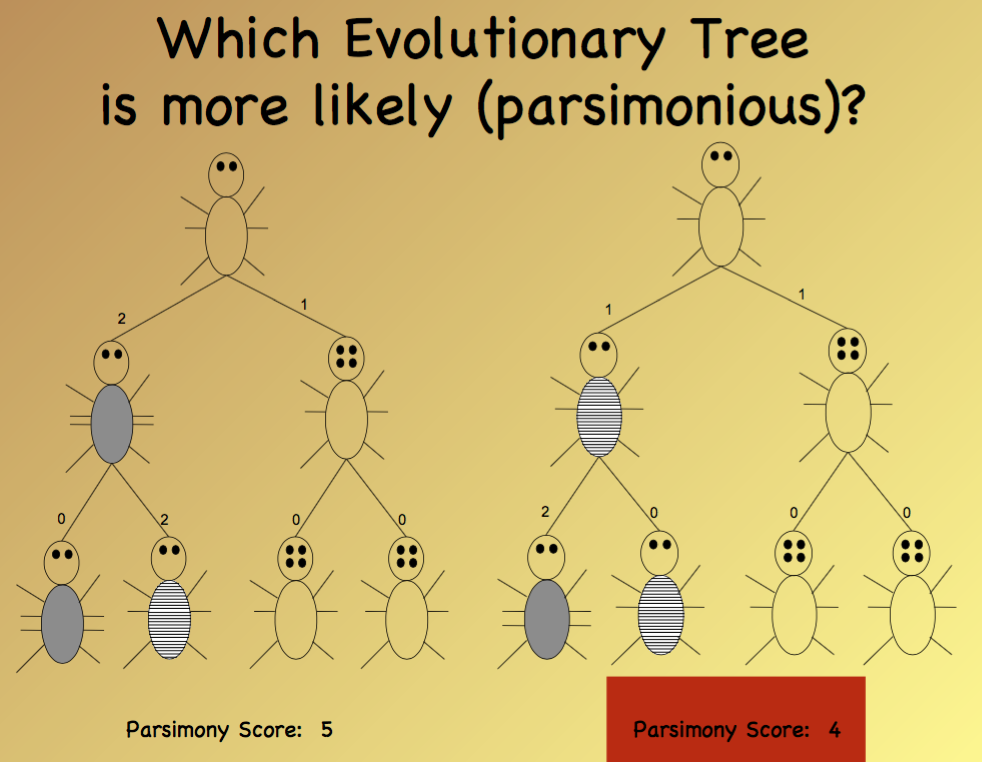
Goal: given an evolutionary "tree skeleton" for several orgnisms, determine the evolutionary change necesary to explain the evolution of the organisms.
We call this problem the small parsimony problem
Parsimonious: adj. unwilling to spend money or use resources: stingy of frugal.
Occam's Razors
In science, we often follow the principle of Occam's Razor:
When competing hypotheses are equal in other respects, the Occam's razor principle recommends the selection of a hypothesis that introduces fewest assumptions while still sufficiently answering the question.
Find most parsimonious ancetors for this tree topology!!
Fitch's Algorithm (Walter fitch)
- Step 1: Do a postorder traversal of the tree and at the same time label every node in the tree with a set of possible best features.
-
Step 2: Do a preorder taversal of the tree and at the same time label every node in the tree with a set of possible best feautres.
-
Traverse tree in postorder
- Leaves:
- Label with the organism's features
- Ancestors:
- If the features of children intersect, label with intersection { this is the set of best features}
- Else label with union of children's features {any feature in union could be ok}
- Leaves:
- Traverse tree in preorder
- Root:
- Pick any labelled feature arbitrarily
- Others:
- Pick label that matches parent feature
- If none, pick any arbitrarily else label with union of children's features
- Root:
Parsimony Problems
- Small Parsimony problem (as above)
- Tree topology is known
- Goal: How many evolutionary changes did occur?
- Solvable in polynomial time (e.g., Fitch’s algorithm)
- Large Parsimony problem
- Tree topology is unknown
- Goal: Find Most Parsimonious Tree
- For this problem no efficient algorithm is known (the problem is shown to b NP-complete)
--
P = NP?
ECS 504 (ustege@uvic.ca)